Next Generation Sequencing (NGS)
What it is:
Reading genetic material faster and cheaper
Sequencing DNA is reading the string of “letters” in the genetic code: the bases A, C, G, and T. Since the 1980s we’ve used Sanger sequencing to read up to 1,000 bases of DNA at a time. We still use Sanger sequencing today, but in the early 2000s, a wave of new DNA sequencing technologies began to emerge that allow us to read hundreds of thousands, or even millions of bases at a time, faster and cheaper than ever before. We can now sequence entire chromosomes, or even entire genomes billions of bases in length, in a single run. We call these technologies Next Generation Sequencing, or NGS.
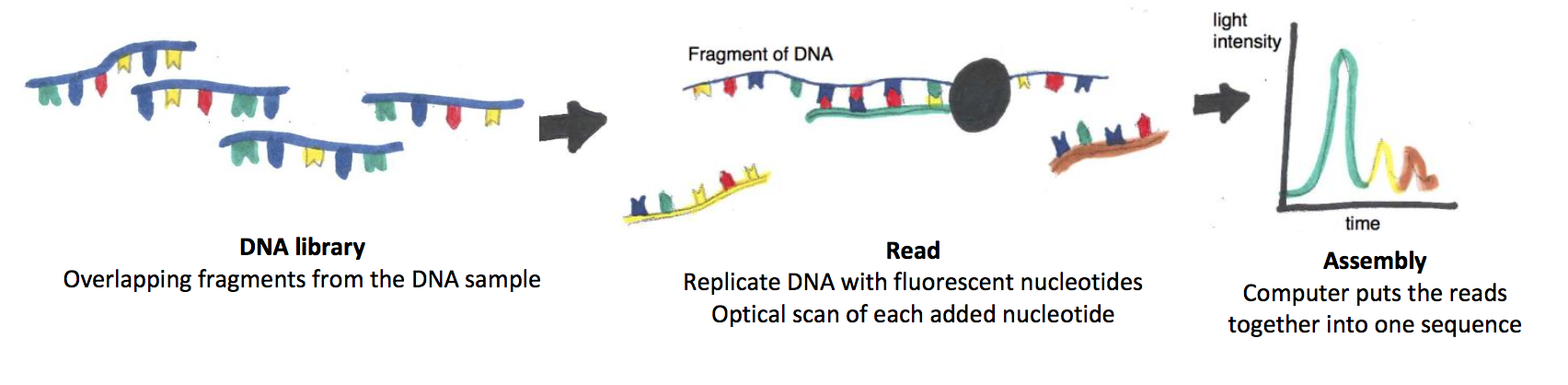
There are several NGS technologies and each works a bit differently, but they all share one thing in common: instead of sequencing stretches of DNA one at a time, NGS can read many different pieces of DNA simultaneously, and then put them back together like pieces of a puzzle. The first step in NGS is to shred the DNA sample that we want to read (say, a two-million base-pair bacterial chromosome) randomly into smaller fragments, to create a DNA library. Fragments in the DNA library will have overlaps, as each of the starting DNA molecules (in our case, many copies of a bacterial chromosome) randomly break at different points in the nucleotide chain. The second step in NGS is to read each of the smaller DNA fragments in the library in parallel with the others. In today’s prevailing NGS technology, the fragments are arrayed onto a small glass slide such that individual library fragments attach to discrete spots on the slide; in our example, the bacterial chromosome is now represented by many thousands of smaller DNA spots, each containing a unique fragment. In each spot, a DNA polymerase (an enzyme that copies DNA) replicates the library fragment by adding complementary nucleotides, where the As, Cs, Gs, and Ts are fluorescently labeled each with a different color. As the DNA replicates, an optical beam scans the slide, and reads which fluorescent color (i.e. which base) is being incorporated into the nucleotide chain for each spot. As a third step, a computer program puts together, or assembles, the overall sequence of the starting sample (our bacterial chromosome) by identifying areas where the sequences overlap. Assembling the overall sequence is easier when the small fragment sequences are aligned to a known sample used as a scaffold (e.g. the full sequence of a similar bacterial chromosome.)
In other words, Next Generation Sequencing works by breaking DNA into smaller pieces to make each piece more manageable to read, then putting those pieces back together to read a complete sequence. The starting DNA sample can be very large – even as large as the entire human genome!
How it is used:
Personalized medicine, gene expression, species ID, and more…
Technology used to sequence the human genome, to read an individual’s entire genetic material, has improved exponentially in the past 15 years. The Human Genome Project took almost 15 years and $3 billion dollars to sequence a single entire human genome. Today, modern technologies can sequence up to 45 human genomes a day for about $1,000 each. A single sequencer today can read many billion bases of DNA per experiment!
This ability to quickly read genetic information has radically changed how scientists and clinicians use sequencing technologies. For example, doctors can now read the entire genome of a cancerous tumor, to help decide which treatment will benefit an individual patient. Scientists can also use NGS to read RNA, to understand when genes are being expressed (turned on or off). We can also sequence very complex samples, such as a microbial swab, to identify many different species present in food, water, and the environment −this field is called metagenomics. Most importantly, Next Generation Sequencing has opened up exciting new clinical applications. The ability to read the genome quickly opens the door to personalized (or precision) medicine, medical treatment that is tailored to an individual’s genetic risks and drug responses.
The future:
Medical and public health improvements
As precision medicine develops further, patients will have access to their own genetic information which they can use to inform medical choices throughout their lifetime. For example, personalized medicine is already being used for more accurate diagnosis, prognosis, and treatment decisions for some cancers. This can also help a patient understand the genetic causes of a previously undiagnosed condition. Sequencing DNA faster than ever before can also be used to discover a genetic basis for ailments for which we don’t understand the cause today.
Researchers also use sequencing technologies to read the entire genomes of infectious agents so they can track the evolution of infectious outbreaks, sometimes with the goal of deploying more effective vaccines against the infection. Scientists can also use sequencing to identify different species by comparing them to a database of known DNA sequences (see the upcoming DNAdot on DNA barcoding!).
The increasing accessibility of whole genome sequencing also means that we may all be able to track our own genomes for new mutations that may appear in the DNA of specific cells during our lifetimes!
Questions – Coming soon!
