Synthetic life: The minimal genome
What it is:
Creating life from scratch?
Creating life where none previously existed has been the stuff of science fiction since Victor Frankenstein. The field of synthetic biology tries to make designing original life forms a scientific reality. Scientists designing synthetic life have several interests, ranging from studying life’s origins to engineering new approaches to medical or industrial problems. Far from victor Frankenstein’s monster however, the life that scientists are trying synthesize today are single-celled bacteria living in petri dishes.
Synthetic biologists take an engineering-like approach to creating cells. By tinkering with the cell’s genetic information and designing new genetic circuits, the hope is to design cells that can deliver new products or perform new processes. A first step toward engineering biological pathways that operate efficiently and cause minimal disruption to cell physiology, however, is to gain a deeper understanding of how a normal cell processes genetic information and how much of that information is redundant, a byproduct of its evolutionary history. Craig Venter and his team have set out to master these challenges by attempting to synthesize the minimal genome – the bare minimum amount of genetic information an organism can use to survive. In 2016 Craig Venter and team announced that they had created an independent organism with the smallest genome yet known. The DNA from this organism was synthesized by a machine, assembled by scientists and then used as the genetic material of a living reproducing bacterial cell. The organism functions with just 473 genes and demonstrates significant advances in the goal to develop life from scratch.
How it works:
Machine-made DNA directing a living cell
The process starts by synthesizing sequences that are roughly 1000 base pairs long. Each end of each sequence is designed to be identical to the end of one other sequence. Several different sequences with these overlapping segments are then put together in a tube with several enzymes. The enzymes match and join the overlapping sequences and the several 1000 base 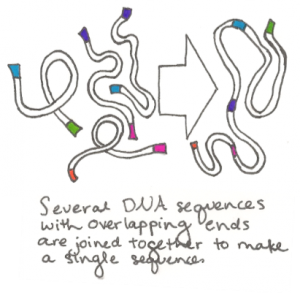 pair segments thus become one single long sequence. Scientists then repeat the process, taking the assembled DNA, now about 10,000 base pairs long, and joining it in the same way with other assemblies they have made. The process continues until the entire genome, all of the original small synthesized segments, has been joined together. This method of joining multiple DNA segments together simultaneously was invented by Daniel Gibson, a scientist at the J Craig Venter Institute, and has since been known as the Gibson assembly method. The assembled genome is then transferred into a related cell that has had all of its DNA removed.
pair segments thus become one single long sequence. Scientists then repeat the process, taking the assembled DNA, now about 10,000 base pairs long, and joining it in the same way with other assemblies they have made. The process continues until the entire genome, all of the original small synthesized segments, has been joined together. This method of joining multiple DNA segments together simultaneously was invented by Daniel Gibson, a scientist at the J Craig Venter Institute, and has since been known as the Gibson assembly method. The assembled genome is then transferred into a related cell that has had all of its DNA removed.
In 2010, using the Gibson assembly method, the team created a JCVI-syn1.0 (short for J. Craig Venter Institute synthetic cell 1.0), a living cell containing a synthesized genome copied from the bacterium Mycoplasma mycoides. As the first living cell with a fully synthesized genome, syn1.0 represented a significant technical achievement. But with about 900 genes and over 1,000,000 base pairs, the genome was simply a slightly modified copy of a naturally occurring genome and was far from minimal.
To create a minimal genome, Venter’s team took the already small Mycoplasma mycoides genome and asked, what can we get rid of and still have a functioning cell? Venter’s team undertook a process of trial and error. The team mixed and matched synthesized segments of DNA, each time identifying whether genes and regulatory regions were necessary for the cell to be viable. In each round of testing, new genes were left out until they arrived at the 473 gene, 531,000 base pair genome that they could pare down no more. This cell they named JCVI-3.0. Their mix-and-match approach allowed them to eliminate not only unnecessary genes, but also unnecessary non-coding regions of the DNA. Perhaps most intriguing about the final genome is that over 31.5% of the genes, 149 in total, are of unknown function.
The future:
A new frontier, or just some new techniques?
There has been some debate on whether JCVI-3.0 actually qualifies as a truly synthetic life form. While the genome that the Venter team created was chemically synthesized, the cell hosting that genome was not. Also, while the synthetic genome was minimized and highly engineered, it still only contained the genes from a single naturally occurring organism. And while the finding that 31.5% of genes necessary for life have an unknown function is biologically interesting, some people question how well something can be engineered if you don’t know how 31.5% of the components work. Still, the creation of the syn3.0 cell was unquestionably a significant technical accomplishment which has advanced the field of synthetic biology in several ways. The Gibson assembly method, used to assemble the various DNA fragments, for example, is already a widely used protocol in the molecular biology field, and the ability to synthesize larger sequences of DNA more cheaply also has widespread implications.
It is still unclear whether synthetic genomes will have a role in drug production or the creation  of biofuels in the future. Now that Venter’s team has demonstrated the ability to completely synthesize a genome, the next immediate questions become how that genome can be modified and added to. Many people believe that new efficient genetic engineering tools such as CRISPR/Cas9 have made the need for creating synthetic genomes unnecessary. Regardless of whether completely synthetic cells become commonplace in the biotechnology of the future, the techniques developed to make the syn3.0 cell have already increased the tools available to biologists, and you can bet that those tools will be used.
of biofuels in the future. Now that Venter’s team has demonstrated the ability to completely synthesize a genome, the next immediate questions become how that genome can be modified and added to. Many people believe that new efficient genetic engineering tools such as CRISPR/Cas9 have made the need for creating synthetic genomes unnecessary. Regardless of whether completely synthetic cells become commonplace in the biotechnology of the future, the techniques developed to make the syn3.0 cell have already increased the tools available to biologists, and you can bet that those tools will be used.
Questions
Review:
- E. coli K-12, probably the most studied bacterium in the laboratory, has more than 4,000 genes. Compare the number of genes in E. coli to the number of genes in the synthetic cell created by Craig Venter in 2016.
- JCVI syn1.0 had roughly 1,000,000 base pairs. Based on the Gibson assembly method, how many original DNA sequences did the team have to synthesize and assemble?
- What was different between the JCVI syn1.0 and syn3.0 cells?
- What are some critiques of the JCVI syn3.0 cell?
- What scientific advance are many other researchers now using that was created by the JCVI syn1.0 and 3.0 team?
Critical thinking:
- Mycoplasma mycoides, the bacterium whose genome was used as the basis for JCVI 1.0 and 3.0 is a parasitic bacterium that infects cattle. Craig Venter’s team originally started working with a cell with an even smaller genome, Mycoplasma genitalium, a sexually transmitted bacterium in humans. These bacteria follow a general rule that parasitic bacteria tend to have much smaller genomes than free living bacteria. Why might bacteria that are parasites have smaller genomes?
- The Gibson assembly method used to piece together JCVI syn3.0 can join up to 10 sequences in one reaction. Knowing that the original synthesized sequences were about 1,000 base pairs each, how many rounds of reactions would it take to make the JCVI syn3.0 cell?
Discussion:
- When JCVI syn1.0 was announced, it led to some controversy. President Obama tasked his Bioethics Commission with studying the possible benefits and risks of such research and developing appropriate guidelines. While genetic engineering of cells, the changing of DNA sequences in living cells, has been done for some time, some think that the synthesis of whole genomes crosses a new line. What guidelines should be placed on the creation of synthetic organisms? Is creating synthetic genomes in bacteria acceptable because of the possible future medical and industrial applications? What about developing synthetic genomes in other organisms, even animals?
Answer key: Available to teachers upon request: dnadots@minipcr.com
