Directed evolution
What it is:
Let biology do the engineering
Imagine being asked to improve an incredibly complex machine, one so complex that you don’t completely understand its inner workings. You would have to start by tinkering with some trial and error. But the chance of succeeding with this approach would be extremely low unless you were able to make a lot of tries and deal with a whole lot of errors—so many that trying them one at a time would be unlikely to get you far. This is the problem that directed evolution tries to solve. The machines to be improved are typically enzymes—proteins that speed up chemical reactions—and the trial and error process uses the principles of evolution by natural selection to tinker with their function.
Scientists would often like to harness the catalytic power of enzymes to perform new chemical reactions, for example to produce new drugs or biofuels. The catch is that designing an enzyme to carry out a specific reaction is very hard. Enzymes are complex three-dimensional molecular machines, and even the most powerful computer algorithms have trouble predicting how changes to a protein’s structure will affect its function. The idea behind directed evolution is that if we don’t have the tools to design an enzyme to do the job we want, maybe we can use the power of natural selection to shape it for us. Using this approach scientists have, in effect, invented an engineering process that requires no knowledge of the structure you are trying to engineer! The approach has been extremely successful, and in 2018 Dr. Frances Arnold was awarded the Nobel Prize in Chemistry for her pioneering work in the field.
How it works:
Mutation followed by selection
The process is called directed evolution because it uses the principles of evolution by natural selection. But unlike evolution by natural selection, directed evolution starts with a definite goal in mind and a person directing the process towards that goal. Both forms of evolution rely on two principles: genetic variation and selection. In nature, genetic variation typically comes from random mutations to the DNA sequence that persist in a population; in directed evolution, the mutations can be more targeted and induced by scientists so that they occur at a very high rate. In nature, selection is determined by which organisms are able to most contribute their genetic information to the next generation; in directed evolution, a selective screen is performed by a scientist who chooses which organisms to continue culturing.
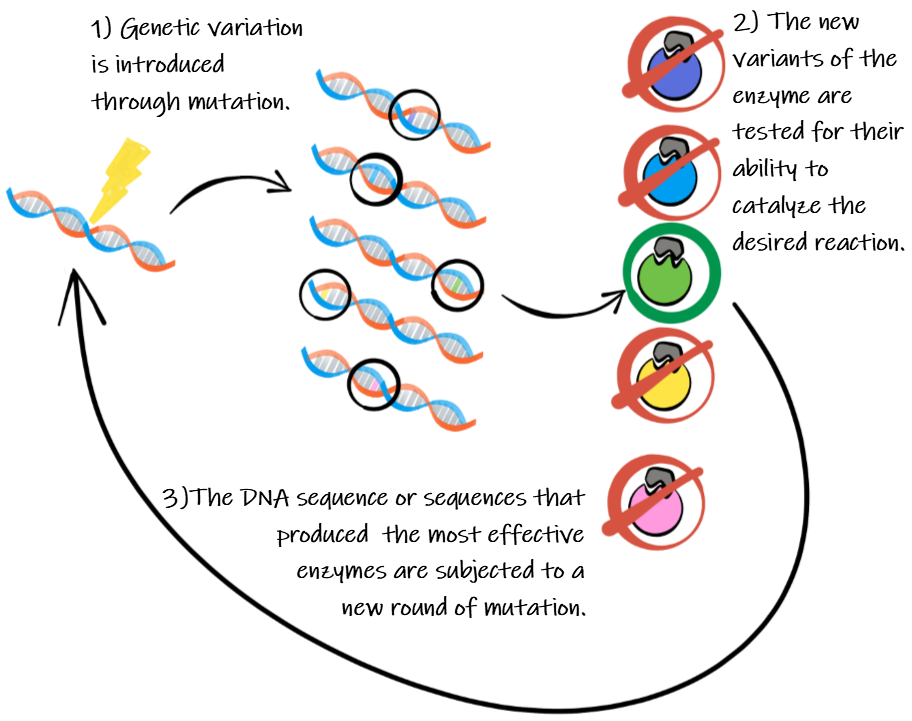
Scientists usually start by identifying an enzyme that performs a reaction similar to what they are interested in and then introducing random mutations to the gene that codes for that enzyme. This results in a library of gene variants that can be tested. Mutations can be introduced in a variety of ways. Chemicals can introduce random mutations across the entire gene. Alternatively, a process known as error prone PCR can introduce random mutations to a targeted area of the gene. Or, recombination could combine sections of different genes together. And while a scientist may not know the exact amino acid changes that will result in the changes they are interested in, often they know enough about the enzyme to know in which areas of the gene changes are more likely to be fruitful and which areas to stay away from, greatly reducing the number of variants that need to be tested. New approaches using more sophisticated computer algorithms have allowed scientists to further narrow their focus when introducing mutations, making the process even more efficient.
Once a library of gene variants has been created, the different enzymes produced by those variants must be tested. Each mutated version of the DNA is placed in a different bacterial strain to express the new mutated form of the enzyme. The scientist will then develop a screen that measures the function of the different forms of the enzyme. Importantly, to be effective this screen needs to be high throughput, allowing the scientist to measure many mutant variants quickly. Most of the new enzymes will likely show reduced function or even no change, but rare variants may show improved function, even if just slightly. The scientist will then take the best performing strains and repeat the entire process, introducing new mutations and then again screening for better enzymatic performance. Every round of mutation and selection should move the enzyme closer towards the goal. In the end, they will have achieved a reengineered enzyme without ever needing to know in advance anything about what changes needed to be made.
Applications, future:
New industrial enzymes and understanding the process of evolution
In many industrial applications, scientists would like to find ways to create molecules efficiently with little or no hazardous byproducts. Creating enzymes to catalyze reactions can be the solution. For example, a major avenue pursued by Dr. Arnold is to develop new ways of converting plants into usable fuels. Plants hold all the raw materials for producing hydrocarbon fuel, such as gasoline, but creating fuels from plants can be inefficient because the enzymes to convert a plant’s cellulose to fuel don’t exist in nature. Through directed evolution, now they do, and researchers are working to place all the enzymes necessary into a single metabolic pathway in one organism, potentially making sustainable biofuels cheaper and more easily produced than ever before.
A large hurdle to effective directed evolution experiments is creating efficient selective screens. The more genetic variants that can be rapidly and efficiently screened, the faster a directed evolution experiment can proceed toward its goal and the more different sequence changes can be explored. New technology will almost certainly make this step of the process easier in the future. And while directed evolution to date has tended to focus on single enzymes, people are now starting to work on evolving whole metabolic pathways, and in the future, it is likely that the process could even be used to create whole synthetic organisms. As these new techniques are implemented, one thing is certain, the world of directed evolution will be continually changing.
Questions
Review:
- In what key ways does directed evolution differ from a normal engineering process?
- In what ways is directed evolution similar to evolution by natural selection?
- In what ways is directed evolution different than evolution by natural selection observed in nature?
- Why do you think being able to do “high throughput” screens would be important in directed evolution?
- People use corn to make ethanol which can be used as fuel. How are scientists looking to make this more efficient?
Critical thinking:
- Imagine if two scientists performed the same directed evolution experiment, in two different labs, completely separately. Do you think they would end with the exact same enzyme at the end of the process? Explain why or why not.
- Why do you think there are some chemical reactions for which no enzymes exist that are able to catalyze the reaction?
- Often when pursuing a directed evolution approach, scientists will choose a few different high performing enzymes to start the next round of mutagenesis, instead of just the best one. What advantage may this have in producing the best enzyme in the end? Why not just choose the single best performing enzyme each round?
Discussion:
- Could the principles of directed evolution be used for other, non-biological engineering problems? What characteristics would the system need to have to make it work? What would be the challenges in trying to use these principles for other engineering problems?
Answer key:
- Available to teachers upon request: dnadots@minipcr.com
