DNA fingerprinting
What it is:
Using DNA variation to identify individuals
Like your fingerprints, your DNA is unique to you, doesn’t change throughout your lifetime, and whether you intend it or not, is often left behind wherever you go. DNA fingerprinting is a method for finding unique patterns in a person’s DNA and using those patterns to identify people, similar to how your unique fingerprint patterns can identify you. But of course, obtaining a DNA fingerprint is a far different process than getting someone’s actual fingerprints. To use DNA as an identification tool, researchers must first obtain quality DNA samples, know where in the DNA to look for identifying differences, and create an efficient, consistent way of looking.
Researchers first came up with a way to do this in the mid-1980s, and more efficient and precise methods have been developed since. To identify differences in 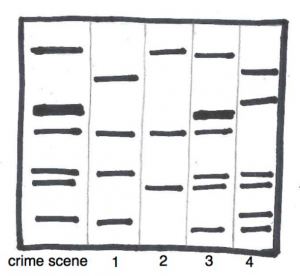 people, researchers look at regions of DNA that are known to vary from person to person. Many of these differences are in regions of DNA that contain repeated sequences known as microsatellites. The most classic protocol to visualize these differences gives the distinctive banding pattern that you have probably seen in crime TV shows and that is illustrated here. To find a match, whether it be for a paternity test or to solve a crime, an investigator simply looks for matching patterns. For example, can you tell which suspect (1,2,3 or 4) matches sample from the crime scene?
people, researchers look at regions of DNA that are known to vary from person to person. Many of these differences are in regions of DNA that contain repeated sequences known as microsatellites. The most classic protocol to visualize these differences gives the distinctive banding pattern that you have probably seen in crime TV shows and that is illustrated here. To find a match, whether it be for a paternity test or to solve a crime, an investigator simply looks for matching patterns. For example, can you tell which suspect (1,2,3 or 4) matches sample from the crime scene?
How it works:
Looking at repeat variations
The variable regions most often used in DNA fingerprinting are specific DNA segments that contain repeated strings of nucleobases. Many places in your DNA, there are regions where the same core sequence of DNA nucleotides is repeated several times over and over again, but the exact number of times the sequence is repeated can vary between individuals. When the length of the individual repeated sequence is short, usually less than 6 base pairs, these segments are called short tandem repeats or STRs (also called microsatellites). Repeats made of longer core sequences are referred to as variable nucleotide tandem repeats or VNTRs (also called minisatellites). In an STR or VNTR, the repeated sequence may occur just a few or even hundreds of times, depending on the specific locus and the individual.
The classic image of a DNA fingerprint comes from looking at VNTRs in a process known as restriction fragment length polymorphism (RFLP) analysis. In this procedure, an investigator takes a DNA sample isolated directly from blood or other tissue and cuts all the DNA in the sample into small pieces using an enzyme known as a restriction enzyme. They then take these DNA fragments and run them on a polyacrylamide gel, which separates all of the DNA fragments by size. Using a process known as southern blotting, the DNA that had been separated by size is then transferred and bound to a nylon membrane. The investigator then washes the membrane with DNA probes. These probes are DNA sequences that match specific VNTRs and will bind to the repeats that have the same sequence. The probes are labeled, usually with a radioactive element, allowing the investigator to create an image of where on the membrane the probes bind. In this approach, the investigator will use several different probes, simultaneously testing several different VNTRs, and so each lane that ran on the gel will contain many visible bands, typically two (one from each chromosome) for every VNTR tested. Because VNTRs are so variable, the chance that two people’s banding patterns on a gel like this will match identically is extremely low. For identifying a suspect from their DNA at a crime scene, you would expect a perfect match for all the bands; for a test like paternity, you would expect that at least half of the bands match between a parent and a child.
RFLP analysis has mostly been replaced by modern methods that are more precise and more sensitive–they can be done with very little DNA input. Today, investigators analyze DNA at a standardized set of more than 20 STRs. Today’s methods typically don’t produce the classic lined patterns that you may be used to seeing in detective TV shows, but they function on the same principle. Unlike classic DNA fingerprints, the investigator will use the polymerase chain reaction (PCR) to make millions of copies of each of the different STR loci. The precise size of each STR is then measured using a method known as capillary electrophoresis. Because the DNA is first amplified by PCR, far less DNA is needed to get a DNA profile. DNA samples can be obtained simply by swabbing surfaces that a person has merely touched, leaving some stray skin cells behind. And capillary electrophoresis is so precise as to tell length differences in DNA down to the single base pair. For criminal investigations, the results of a genetic profile for these different STRs are stored by the FBI in a system known as Combined DNA Index System (CODIS). CODIS is a standardized set of STR markers that all criminal labs test, meaning that no matter who does the DNA analysis, the results can be searched and analyzed in a standard way.
The future:
Technology just keeps on moving
DNA fingerprinting techniques introduced in the 1980s revolutionized forensics technology, and the CODIS system has been extremely effective in identifying criminals and exonerating the wrongly convicted. But CODIS was first implemented in the early 1990s and DNA technology has improved significantly since.
A new frontier in genetic profiling has now opened with the advent of direct to consumer genetics testing. More and more people are voluntarily obtaining their own genetic information and even sharing it online to find long lost relatives. But the same information used to identify the third cousin you didn’t know you had can be used to identify criminals. Police have begun to use these databases, and even if they don’t get a match for their suspect, they can identify family members who have submitted their own DNA, greatly narrowing down their suspect pool. While the CODIS database is constrained to searching among people who have at least been arrested, ancestry genetic profiles contain samples from the general population and contain significantly more genetic information than CODIS. In the case of Golden State Killer, police were able to identify and arrest a suspected serial killer more than 30 years since he last committed a crime, without ever obtaining his DNA directly. Still, there are concerns over privacy. While the arrest of a serial killer is clearly a good thing, some argue that being able to identify virtually any person by merely swabbing a surface they once touched and comparing that DNA to a distant cousin’s online profile goes far beyond the idea of taking a person’s fingerprint.
Learn more:
- “Southern Blotting and DNA Fingerprinting.” Lasker Foundation.
- “Thirty Years of DNA Forensics.” Chemical & Engineering News.
Questions:
Review:
- What are some factors that make DNA especially good for use in identification?
- What makes microsatellites a good place to look for variation?
- When DNA fingerprinting was first introduced it could only be used in a few select cases where a decent amount of blood or other tissue was left at a crime scene. What key innovation made it so that fingerprinting can be used in more and more cases?
- What is similar about both the CODIS, STR method and the more traditional Southern blotting techniques of DNA fingerprinting?
- In what ways is looking at publicly available genetic ancestry data different from more traditional DNA fingerprinting technique?
Critical thinking:
- In a traditional gel-based DNA fingerprint for a perfect match, you would expect all the bands on the gel to match. What percentage of the bands do you think you would expect to match for two siblings? A grandparent and grandchild? Explain why you think that.
- In some cases, at a crime scene, blood or other tissue from two people is mixed. Could you still use this for identification? If not, explain why. If so, explain how.
Discussion:
- Police are now testing public databases to look at individual’s DNA and try to catch criminals. Without laws dictating how police access this information they will likely continue doing so more and more. A lot of people who submit their DNA to these sites are just doing it for fun, looking for long lost relatives and have no idea their DNA could be used in this way. Is it OK for police to use this genetic information without getting approval from individuals first? Does it make you more or less likely to want to have your own DNA tested?
Answer key:
Available to teachers upon request: dnadots@minipcr.com
