Nanopore sequencing
What it is:
Sequencing without base pairing
The goal of DNA sequencing is to determine the exact order of nucleotides, adenine (A), thymine (T), guanine (G), and cytosine (C), in a DNA molecule. The difficulty is that DNA is far too small to visualize directly. Since DNA sequencing was first invented in the 1970s, every sequencing method has relied on the rules of complementary base pairing to determine the order of nucleotides. By allowing other nucleotides to bind to the sequence we are interested in, and somehow keeping track of what nucleotides were used in that process, we can determine our original sequence. In contrast, nanopore sequencing is able to sequence a strand of DNA or RNA directly, reading the nucleotides one by one, without the use of base paring rules.
To understand how nanopore sequencing works, it helps to think of an old film movie projector. The film itself is a very long wound up strand made of individual pictures ordered in a very specific sequence. To see each picture, the film is pulled through the projector. As each picture enters the projector, a flash of light passes through, allowing us to see the images, one by one. In nanopore sequencing, the nanopore works much like the film projector. A long strand of DNA is pulled through the nanopore, but instead of using a beam of light, a current of charged ions is used to determine the very specific sequence, one nucleotide at a time, as the strand passes through.
How it works:
Measuring changes in electric current
A nanopore is a hole in a material that is approximately a nanometer in diameter, a little bit wider than a single strand of DNA. In nature, often the only way to get across a cell membrane is to travel through a pore created by a protein, and there are many, many different types of proteins that allow different types of molecules to pass through. In nanopore sequencing, an electric current is passed across a synthetic membrane that contains a protein nanopore. The membrane is resistant to the electric current, meaning the only way for current to flow across the membrane is to travel through the pore. Because DNA and RNA are charged molecules, the electric current also pulls DNA and RNA through the pore with it, in the same way a current pulls DNA or RNA through agarose in gel electrophoresis. The pore, however, is only large enough for a single strand to fit through, one nucleotide at a time. Because the nanopore is so small, when a nucleotide is in the pore, current passing through the pore is partially blocked, each nucleotide blocking the current in a distinctive way. By measuring the change in current caused by the nucleotide, scientists can use algorithms to figure out which nucleotide caused the change. This is much like how the light passing through a projector is blocked by each frame of film in a distinctive way, allowing us to see the picture. 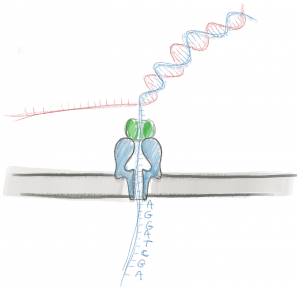
To sequence a DNA or RNA sample, the end of each DNA or RNA molecule is first attached to a “sequencing adapter”, a small piece of DNA with an enzyme motor wrapped around it. The sample is then put on the nanopore membrane. When an electric voltage is applied to the top and bottom of the membrane, the DNA is pulled into the nanopore until the enzyme motor sits on the nanopore like an oversized cork that is too big to fit into a bottle, and the strand of DNA dangles into the nanopore. The enzyme motor will then work as a helicase, unwinding the DNA double helix. As the DNA is unwound, the enzyme motor allows a single strand of nucleotides to pass through the nanopore at a rate that is slow enough to measure the change in ionic current, nucleotide by nucleotide, until the entire strand has passed through. This process allows for very long sequences to be read. Where other sequencing technologies typically only read a few hundred nucleotides at a time, nanopore sequencing can regularly read sequences that are tens of thousands of nucleotides long. These long sequence reads can be a huge advantage when repetitive elements and other types of sequences can make it difficult to assemble many short sequences into one long one. Once the entire nucleotide strand passes through the pore, a new nucleotide strand will be pulled in, and the nanopore will start sequencing again. And because nanopores are so small, thousands of nanopore membranes can fit on a grid the size of a postage stamp, each simultaneously sequencing a different strand from the same original sample.
The future:
Better, cheaper sequencing in a pocket-sized machine
Nanopore sequencing does have its disadvantages. It tends to be error prone; error rates in nanopore sequencing can be as high as 15%. If sequencing large amounts of the same sequence, this error rate can be tolerable because multiple copies of the same sequence will allow the user to recognize and eliminate mistakes. When looking to study small or rare sequence differences, however, such error rates can be a big problem. Changes to the proteins involved in the sequencing process have dramatically reduced this error rate over time, and in the future, it is only expected to become more accurate. And while nanopore sequencing is around the same cost as other sequencing technologies, current nanopore membranes have a limited shelf life before they need to be replaced at a cost of several hundred dollars. So-called solid-state nanopores that are in development will no longer use protein channels, instead using pores made directly in synthetic materials. This could increase the lifespan of the membranes, allowing membranes to be stored for much longer periods of time, and potentially fitting many, many more nanopores into an array, reducing the cost of sequencing even further.
A major advantage to nanopore sequencing machines is that they can be very small, use few reagents, and need very little power. While other sequencing technologies use machines that can be the size of a dishwasher, current nanopore sequencers can easily fit in your pocket and are powered by a USB connection to a laptop computer. Scientists are now bringing mobile laboratories to previously inaccessible places, collecting organisms and sequencing their DNA, all without leaving the field, and in 2016, the International Space Station added nanopore sequencing to its scientific capabilities. What and where scientists will sequence next is anybody’s guess, but we do know this: it is likely to be where no sequencer has gone before.
Questions:
Review
- How is nanopore sequencing different than other more traditional sequencing techniques?
- In nanopore sequencing, what passes through the nanopore?
- What does the nanopore sequencer actually measure to determine the nucleotide sequence?
- Why must a protein motor be added to the end of the nucleotide sample before putting on the nanopore membrane?
- Why could solid state nanopores reduce the cost of nanopore sequencing in the future?
Critical thinking
- Nanopore sequencing is favored by many scientists for its long sequence reads. Why would DNA with long sections of repetitive sequences be more difficult to sequence using more traditional sequencing techniques?
- If solid-state nanopores could produce pores that have a longer life span, what could be some reasons that scientists made protein based nanopores first?
Discussion
- As DNA sequencing continues to become cheaper and cheaper and easier and easier, we are moving into a world where being able to pay to have your own entire genome sequenced is becoming a real possibility. If you could get your own genome sequenced, would you? What would be some of the advantages? The downsides?
Answers available to teachers upon request. Email dnadots@minipcr.com.
