Quantitative Polymerase Chain Reaction (qPCR)
What it is:
Monitoring how many copies of DNA are produced by PCR
If you go to the finish line of a race after the race is over, there is some information you can learn, but some you cannot. You can tell who ran the race, but not who won; the winners and losers will all be there standing around together. If you want to know who wins, you need to watch the race as it happens. The same is true for PCR.
PCR, or polymerase chain reaction, is one of the most common and powerful techniques in a molecular biology laboratory. PCR gives scientists the ability to find a specific sequence of DNA in a complex sample and make billions of copies of that specific sequence, all in a couple of hours. But while PCR is an incredibly powerful tool, it is typically an end-point reaction, meaning once we start a PCR, we don’t look at it again until the reaction is over – we don’t watch as the reactions cross the finish line. Quantitative PCR (qPCR), also known as real-time PCR, puts a spin on this process by monitoring the reaction in real time using fluorescence to label the copies of DNA as they are produced. During qPCR, the amount of fluorescence that is measured is directly proportional to the amount of DNA being produced – the brighter it glows, the more DNA that is present. As each new copy of DNA produced is fluorescently labeled, the reaction will fluoresce a little brighter, allowing researchers to keep quantitative track of how much DNA is produced during each cycle of PCR. By tracking the rate of the reaction, and comparing that rate to other reactions, it’s possible to calculate relatively how much of your very specific DNA sequence you started with.
How it works:
Tracking the path of PCR
In each cycle of a PCR, the amount of the specific DNA sequence you are amplifying should theoretically double, leading to exponential growth. Early in the reaction there will not be enough target DNA and the rate of copying will be relatively slow, but as the DNA doubles every cycle, eventually the reaction starts to fluoresce brighter and brighter, faster and faster. Eventually, this growth will level off and reactions will all appear the same brightness as they become saturated. Earlier in the process, however, very clear differences can be observed, and how many cycles it takes a particular reaction to fluoresce brightly enough to be detected
above background levels can be measured very consistently. A qPCR machine will create a graph where the fluorescence from each individual reaction can be monitored in real time. The point at which fluorescence is first measured above background levels – where the curve is first registered reliably on the graph – is called the Ct, or the “threshold cycle”. The sooner Ct is reached, the more template DNA was in the tube to start with.
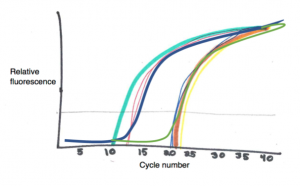
What qPCR does not do is give an absolute count of DNA molecules. qPCR is able to quantify DNA only relative to other reactions, so it is very good at comparing amounts of DNA between two samples. As long as the DNA input differs by at least two-fold, very accurate relative concentrations of starting template can be established. To get an absolute count of starting molecules a scientist must also run a standard, which has a known starting DNA concentration. This way, through comparison, a more exact estimation of starting DNA template can be established.
How it is used:
Gene expression and molecular diagnoses
Perhaps qPCR’s biggest contribution has been in the field of gene expression. Often a scientist is interested not in how much DNA is in a sample, but how much of a particular messenger RNA (mRNA) is present. By measuring mRNA levels from particular genes, you can determine how much that particular gene is being used in different tissues or in the same tissue under different conditions. Such studies require a first step of copying all the RNA in a cell into DNA using a process called reverse transcription. The newly transcribed DNA is then run in a qPCR reaction as described above. The reverse transcription step has led this procedure to be known as RT-qPCR, and RT-qPCR has been considered the gold standard for validation of gene expression levels for about two decades.
While qPCR may have made its biggest mark in the field of gene expression, most any time a scientist wants to quantify an amount of DNA, qPCR can be used. Being able to not just detect, but also quantify, has led qPCR to be used more and more in the medical field for diagnoses. By using qPCR, a physician can tell not only if a particular DNA sequence is present, but they can also tell whether it is abundant or rare. For example, often, a physician may be interested in whether a virus is present in the body, as well as in what the patient’s viral load is. qPCR can answer both questions in a single test, and it can be used in diagnosing, HIV, hepatitis, and many viral pathogens. Similarly, qPCR is used for identifying cancers, both recognizing particular mutations in the body and measuring their prevalence. In environmental work, when measuring pathogen load in a sample of food or water, scientists may not just want to know if a particular strain of bacteria is present, but also how widespread it is.
But in molecular biology, no test remains the gold standard forever. New methods of quantifying DNA are already in use and are able to measure even rarer DNA sequences in even greater precision. qPCR will no-doubt be around for a good while longer, but as scientists look to gain more and more sensitivity and precision in their tests, even classic techniques such as PCR continue to be reinvented to meet the new demands.
Questions:
Review:
-
- What does the “q” in qPCR stand for? How does that make qPCR different than regular PCR?
- Regular PCR does not require a fluorescent dye or probe. Why is fluorescence required for qPCR?
- Explain the difference between an end-point and real-time experiment.
- Why must a qPCR machine take data in real time? Why is just taking one reading at the end not useful.
- Why would qPCR be useful in diagnosing disease?
Critical thinking:
-
-
- Consider DNA is amplified in a PCR. Why can qPCR only distinguish between a greater than two-fold difference in starting concentration?
- If one sample reaches Ct four cycles later than another, about how large would you expect the difference in starting concentration of DNA is?
-
Discussion:
-
-
- qPCR has been the considered the “gold standard” for quantifying DNA and RNA for about two decades. But now, some even more precise technologies are being developed. Some people think that qPCR will soon no longer be used, while others think that because of its utility will always have a significant role in biological laboratories. What do you think is more typical; when a new technology is introduced are the old technologies no longer useful and eventually forgotten, or are do both technologies continue to be used together? Can you think of examples, scientific or otherwise, of each case?
-
Answer key: Available to teachers upon request: dnadots@minipcr.com
