Riboswitches
What it is:
RNA – not just a messenger!
For a long time, RNA was viewed mostly as just a simple intermediary between DNA and protein. To make a protein, a DNA sequence, or a gene, must first be copied into messenger RNA (mRNA) in a process called transcription. Next, in a process called translation, the mRNA will be read at the ribosome where amino acids will be joined to form a protein. But not all genes are used all the time. When a gene is used to make a protein, we say that the gene has been expressed. Changes in gene expression affect how cells function, acquire specialized shapes, or respond to environmental stimuli. For a long time, scientists thought that changes in gene expression were regulated almost exclusively at the level of transcription, the production of mRNA from DNA. Today, however, scientists know that changes in gene expression can also occur after the mRNA is made.
In a stunning example of how gene expression can be controlled after mRNA is produced, scientists discovered that some bacteria make specialized mRNAs that can turn themselves on and off. Like all mRNAs, these specialized mRNAs have a coding region which contains the information to make a protein, but they also carry a specialized noncoding region that controls whether or not translation will occur. These specialized noncoding regions work as a switch, turning translation on or off and thereby controlling gene expression. Because these switches are made of ribonucleic acid (RNA), scientists call them riboswitches.
How it works:
RNA molecules that control their own expression
While RNA is often depicted as a linear molecule, RNA molecules can also twist and fold up as the nucleotides bind with each other. Like in DNA, this binding is guided by base paring rules, as adenine (A) binds to uracil (U), and guanine (G) binds to cytosine (C). Riboswitches contain stretches of complementary RNA bases that interact and fold into such three-dimensional shapes. These regions where the RNA binds to itself are known as RNA secondary structures.
Riboswitches contain two specialized secondary structures. The first is called an aptamer. An aptamer is a structure that can bind to a specific small molecule. The specific molecule an aptamer will bind to is called a ligand. The second secondary structure is an expression platform. The expression platform responds when the aptamer and ligand bind and impacts translation of the mRNA at the ribosome. Each riboswitch responds to a different ligand, depending on the specific structure of the aptamer region. When the aptamer region binds its ligand, it triggers structural change in the expression platform. This structural change regulates the coding region of the mRNA by either increasing or decreasing translation. In this way, a riboswitch allows an mRNA molecule to regulate its own expression.
Riboswitches respond to ligands as a mechanism to regulate cell function in a changing environment. Some riboswitches work by increasing gene expression when a ligand becomes more abundant. For example, the crcB riboswitch is part of an mRNA that codes for a fluoride ion transport protein. The crcB aptamer is activated by binding to fluoride ions. When the aptamer binds to fluoride, the expression platform works to increase gene expression, thereby making more fluoride ion transport proteins. This increased production of ion transport proteins then lowers the fluoride ion concentration in the cell to a normal level.
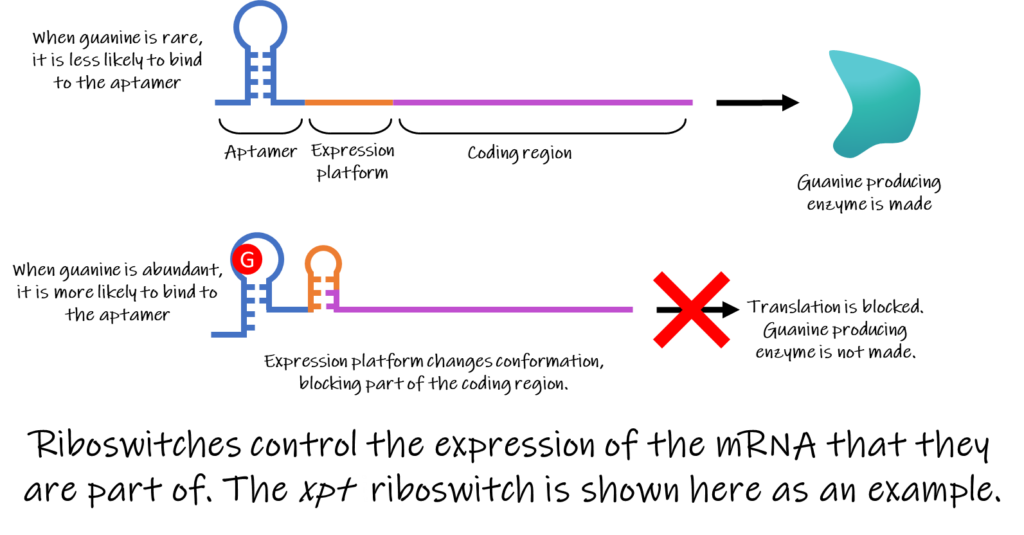
Other riboswitches work by blocking the production of the protein encoded by the mRNA. Some riboswitches achieve this by actively cutting the mRNA in a form of self-destruction when the ligand binds. Others work by simply changing the conformation of the mRNA in a way that blocks the ribosome from being able to translate the mRNA. For example, the xpt riboswitch binds to guanine (see figure), one of the bases used to build DNA and RNA. When guanine is abundant in the cell, it is more likely to bind to the xpt riboswitch. When guanine binds to the aptamer region of the xpt riboswitch, the expression platform changes shape and blocks the translation of an enzyme needed to produce guanine. As less of the enzyme is made, less guanine is created, and guanine levels return to a normal level. When guanine levels are low, the riboswitch is less likely to be activated, and more of the guanine-producing enzyme will be made.
The future:
A new way to control gene expression
Riboswitches were discovered as natural parts of some bacterial gene expression systems, but scientists are applying these naturally occurring riboswitches to new contexts and are also designing new riboswitches that don’t exist in nature. In a lab setting, scientists often look for new ways to control protein expression, either for research purposes or for industrial protein production. Bioengineers can now use riboswitches as a new tool to control the expression of specific proteins that they are interested in.
Other researchers are looking to take advantage of the fact that riboswitches are not naturally present in human cells but are common in many bacterial pathogens. Researchers are developing new ligands, designed to activate existing bacterial riboswitches with the goal of disrupting gene expression and killing the cell. If administered as a drug, these new riboswitch ligands have the potential to act as a new class of antibiotics.
Regardless of whether they are used to kill bacterial cells or control the production of proteins in the lab, riboswitches are finding their way into the molecular biology toolkit. They are also one more example of how RNA has proven to be much more than just a simple messenger.
Questions
Review:
- What is gene regulation is and why it is it important?
- How did riboswitches get their name?
- What are the two important parts that make up a riboswitch? What is the role of each?
- For the xpt riboswitch, explain what happens when the amount of guanine increases in the cell.
- Riboswitches are made of RNA. Why are many protein engineers interested in riboswitches?
Critical thinking:
- In nature, the ligand for a riboswitch is usually directly related to the protein that is encoded by the mRNA. But in the lab, a scientist could engineer a riboswitch into any mRNA. Why would it be important that the ligands for natural riboswitches be directly related to the protein that is encoded by the mRNA? Why might it be important to a scientist to be able to use different riboswitches on different RNAs?
- Negative feedback and positive feedback are important concepts in how cells respond to changes. In negative feedback, a stimulus causes a response that goes on to reduce the original stimulus. In positive feedback, a stimulus causes a response that goes on to increase the original stimulus. Are the riboswitches described in this article examples of positive or negative feedback mechanisms? Explain your reasoning.
Apply what you learned:
- There are lots of types of riboswitches that function in different ways. Use what you learned from this reading and design your own riboswitch! Think about a biological process that you would like to regulate in a cell and identify a key protein and a key small molecule that is involved. Would you want the binding of the small molecule to your riboswitch to cause an increase or decrease in the protein production? What would your expression platform do to achieve your intended impact on protein production? Diagram your riboswitch on paper. Make sure to label the aptamer domain, expression platform, as well as explain how this riboswitch regulates your biological process. Be creative; you’re not creating a real riboswitch, just one that could work in theory.
Answer key:
Available to teachers upon request: dnadots@minipcr.com
