RNA Interference (RNAi)
What it is:
There’s more than one way to turn off a gene
For a cell to develop and function, it is essential that it can turn specific genes on or off at different times. Normally, we think of turning a DNA sequence into a protein as a fairly linear process – what is often referred to as the “central dogma” of molecular biology. DNA is transcribed into messenger RNA (mRNA) in the nucleus, and mRNA is translated to protein at the ribosome. From this simple model, you could think that the way to make one cell different from another is to change what stretch of DNA in the genome is transcribed into mRNA at the start of that process. Indeed, for decades scientists thought that turning transcription on and off was the only way to regulate gene expression. But it turns out that there is a second place where cells can control the flow of information from DNA to protein. The cell can also regulate how much mRNA gets translated into protein. RNA interference (RNAi) operates at this level, by way of tiny RNAs that are complementary to specific mRNAs and that can halt those mRNAs from being decoded into proteins at the ribosome. 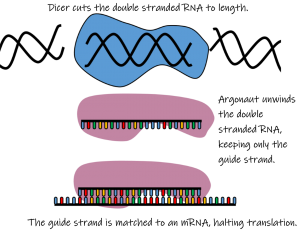
Scientists have known for some time that adding so-called antisense RNA, RNA that is complementary to a specific mRNA sequence, can interfere with that mRNA being transcribed. But in 1998, working with the nematode C. elegans, Craig Mello, Andrew Fire and colleagues showed that adding double stranded RNA, instead of single stranded, made the effect far, far more pronounced, such that just a few double stranded RNA molecules could inhibit virtually all expression of a particular gene. The effect was so strong that they proposed that they had tapped into a previously unknown method of gene regulation. The era of RNAi had begun, and in 2006 Mellow and Fire were awarded the Nobel Prize.
You can think of RNAi like this: there are two ways to not get a letter from a friend. First, your friend could not write the letter. Second, your friend might write the letter, but a thief may snatch it before it arrives at your house. RNAi is like that thief, stealing mRNA sequences before they can be used by the ribosome.
How it works:
Killing with complementarity
The RNAi pathway starts with double-stranded RNA. Just like DNA, two strands of RNA that are complementary to each other can bind together and form a double helix. RNAi works by cutting up double stranded RNAs and then using the cut-up pieces to block the production of protein from mRNA. Different sources of double stranded RNA can serve as a starting point for the RNAi pathway, some internal and some external to the cell. In some organisms, RNAi can work as part of a cell’s defense mechanism; take RNA viruses as an example. The RNAi pathway recognizes the double stranded RNA of the viral genome, cuts it into shorter pieces, and then uses the resulting small RNAs to specifically target and “silence” viral genes, blocking translation of viral proteins. The cut-up short pieces of RNA used in the pathway are referred to as short interference RNAs (siRNA). Cells can also produce their own small double stranded RNAs as part of normal gene regulation. A common form of endogenous (originating from inside the cell) small RNA is known as microRNA (miRNA). miRNAs start as shorter, single-stranded RNA molecules that fold over and bind to themselves making a partial double stranded structure known as a stem-loop. miRNAs are encoded by the cell’s own genome and can track down the cell’s own mRNAs, effectively regulating which ones are allowed to make proteins.
miRNAs and siRNAs function very similarly in how they silence gene expression. Both start their lives as double stranded RNA molecules. After some pre-processing, the protein dicer cuts the double stranded RNA to its final length of approximately 20-30 base pairs. This short, double-stranded RNA next binds to another protein known as argonaute, where it unwinds and becomes two single strands. One of the two strands is released and degraded, while the other, called the guide strand, remains bound to argonaute and goes on to form the RNA-induced silencing complex (RISC.) RISC relies on base pairing rules to identify mRNAs that are complementary to the guide sequence. If it finds a complementary sequence in an mRNA, that mRNA will be blocked from being translated into protein. For siRNAs, this typically leads to the mRNA being cut in two by argonaute; for miRNAs, RISC typically binds in the mRNA 3’ untranslated regions and blocks translation by the ribosome.
Applications:
Gene knockdown to treating disease
The discovery of RNAi immediately gave researchers a new tool to manipulate the inner workings of the cell. By delivering siRNA to a cell, researchers can effectively silence the expression of specific genes, and they can do this in an otherwise intact genome. The more traditional way of investigating a gene’s function is to try to “knock out” the gene through some method of mutagenasis, eliminating the DNA sequence entirely or making it non-functional – a traditionally difficult and costly approach. With RNAi, scientists can take otherwise normal organsims and directly “knock down” the production of proteins derived from a particular gene. And because producing custom RNA sequences is relatively easy and affordable, scientists can now order custom siRNAs to test the effect of silencing virtually any gene. This has proved extremely useful in studying human cell lines grown in laboratory culture, model organisms, such as Drosophila and C. elegans, and even non-model organisms where genetic tools have not been previously available. 
The possibility of knocking down expression of a particular protein has been particularly alluring to the medical field. If doctors could turn off an important gene needed in the growth of cancer tumors, or one responsible for a genetic disorder, diseases could be effectively halted in their tracks. The main problem with this approach is that it is very difficult to get RNA from outside the body into a particular cell. Nucleic acids like RNA typically do not diffuse through cell membranes, and the body is filled with RNAses, enzymes that degrade stray RNAs. But scientists have made progress in packaging siRNAs such that they can now be delivered to some cells effectively. In 2018, a treatment for hereditary transthyretin amyloidosis, a genetic disorder caused by a protein produced in the liver but that progressively affects nervous system and otherwise leads to death, became the first RNAi therapy to receive approval from the FDA. That approval came just 20 years after RNAi was first described in the journal Nature – not a bad anniversary gift.
Questions
Review:
- What are the two basic steps in which gene expression can be regulated?
- What are some differences between siRNA and miRNA?
- Describe what happens to the two strands of double stranded RNA after they are bound to argonaute.
- What advantage might scientists have using RNAi have over traditional gene knockout studies?
- Why would simply taking a pill with siRNA or injecting a patient with siRNA to silence a defective gene not be an effective treatment?
Critical thinking:
- Would you describe RNAi as an invention or a discovery?
- One difference between miRNA and siRNA not mentioned in the text is that siRNA tends to only bind with perfect homology – when all 21 base pairs find their match. miRNAs bind with less specificity, typically only about eight bases need to find their match. What does this say about the number of different mRNA sequences siRNAs are likely to regulate compared to miRNAs?
Discussion:
- If mechanisms to deliver siRNAs to human cells become more reliable and easier to use, discuss what you think some of the advantages of using RNAi could be. What could some uses be? Can you think of genes that you might want to turn off in the body, but only temporarily?
Answer key: Available to teachers upon request: dnadots@minipcr.com
